"Illuminating the active virosphere with BONCAT and single virus genomic sequencing technologies"
https://www.biorxiv.org/content/10.1101/2025.08.05.666878v1?rss=1 #Dynamics #Cell
#dynamics
"Bistability in Gene Regulation: Simulating Positive Feedback and Toggle Circuits Using Python and Hill Functions"
https://www.biorxiv.org/content/10.1101/2025.08.05.668746v1?rss=1 #Dynamics #Cell
"Automated 1D Helmholtz coil design for cell biology: Weak magnetic fields alter cytoskeleton dynamics"
https://doi.org/doi:10.1371/journal.pone.0321133
https://pubmed.ncbi.nlm.nih.gov/40763285/
#Dynamics #Cell
"Structural determinants for GqPCR-mediated inhibition of TASK K2P K+ channels and their dysfunction in disease"
https://www.biorxiv.org/content/10.1101/2025.08.04.668454v1?rss=1 #Dynamics #Cell
"Ex vivo live imaging unveils the dynamics of oocyte formation in mice"
https://www.biorxiv.org/content/10.1101/2025.08.04.668457v1?rss=1 #Dynamics #Mitosis #Cell
"MIDOE: Maximally-informed Design of Experiment to infer experimentally inaccessible transcription factors dynamics"
https://www.biorxiv.org/content/10.1101/2025.08.04.668461v1?rss=1 #Dynamics #Cell
"HES1 oscillations are required for cell cycle progression and re-entry in oestrogen receptor positive breast cancer cells"
https://www.biorxiv.org/content/10.1101/2025.08.04.668440v1?rss=1 #Dynamics #Cell
"A critical role for VE-cadherin in regulating actin dynamics during endothelial maturation and non-inflammatory activation via a tension-sensitive intermediate state"
https://www.biorxiv.org/content/10.1101/2025.08.05.668624v1?rss=1 #Mechanical #Cadherin #Dynamics #Cell
The eleventh day of the Konstanz School of Collective Behaviour 2025 (#KSCB2025) starts with a #keynote by Aneta Koseska on the dynamical basis of natural #computations across biological scales.
"miRNAs and exosomes in psoriasis: coordinating cytoskeleton dynamics and extracellular matrix remodeling"
https://doi.org/doi:10.3389/fcell.2025.1608902
https://pubmed.ncbi.nlm.nih.gov/40756258/
#Cytoskeleton #Cytoskeletal #Dynamics

"Allele-Specific Effects of Mutations in the Rifampin Resistance-Determining Region (RRDR) of RpoB on Physiology and Antibiotic Resistance in Enterococcus faecium"
https://www.biorxiv.org/content/10.1101/2025.07.25.666922v1?rss=1 #Dynamics #Cell
"Physiological Reconstitution of Microtubule Doublets"
https://www.biorxiv.org/content/10.1101/2025.08.03.668368v1?rss=1 #Microtubule #Dynamics
"Hedyotis Diffusae Herba Mitigates Rotenone-Induced Neurotoxicity via Akt Pathway in SH-SY5Y Cells"
https://www.biorxiv.org/content/10.1101/2025.08.03.668347v1?rss=1 #Dynamics #Cell
In our recent #JournalClub, I presented Genkin et al. (2025), who decode #DecisionMaking in the #PremotorCortex of #macaques as low-dimensional #latent #dynamics shared across #NeuralPopulations. Their generative model links tuning curves, spike-time variability, and stimulus-dependent potential landscapes to a common internal decision variable. I summarized and discussed their findings in this blog post:
https://doi.org/10.1038/s41586-025-09199-1
https://www.fabriziomusacchio.com/blog/2025-08-01-decoding_decision_making_in_premotor_cortex/
"Rapid Single-Cell Measurement of Transient Transmembrane Water Flow under Osmotic Gradient"
https://arxiv.org/abs/2508.00104 #Physics.Bio-Ph #Dynamics #Cell

"Dynamics of a Data-Driven Low-Dimensional Model of Turbulent Minimal Pipe Flow"
https://arxiv.org/abs/2408.03135 #Physics.Flu-Dyn #Dynamics #Cell

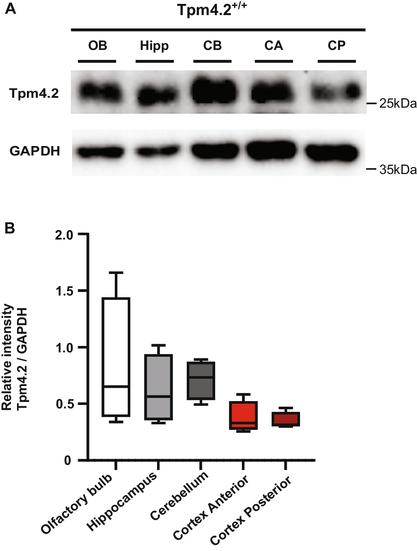
"Knock-out of Tpm4.2/Actin Filaments Alters Neuronal Signaling, Neurite Outgrowth, and Behavioral Phenotypes in Mice"
https://doi.org/doi:10.1007/s12035-025-05259-9
https://pubmed.ncbi.nlm.nih.gov/40753314/
#Dynamics #Actin
"TRPM7 Annexin A1 Mechanosensitive Pathway Drives Capillary Infiltration by Circulating Tumor Cells"
https://www.biorxiv.org/content/10.1101/2025.08.01.668151v1?rss=1 #Dynamics #Actin


